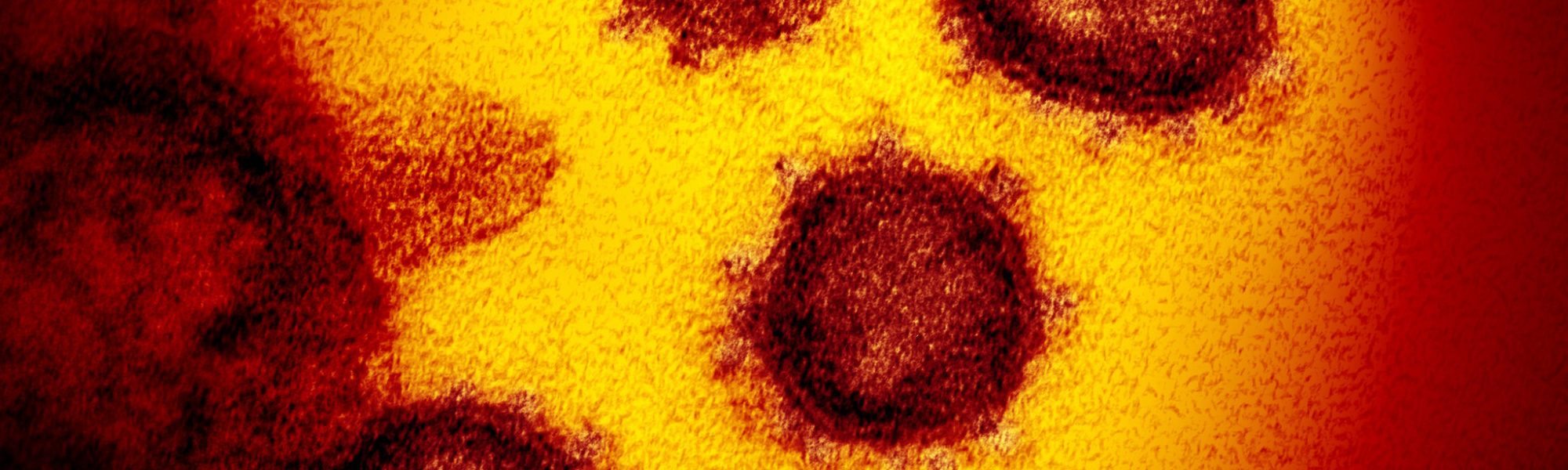
A detailed look inside SARS-CoV-2, the virus responsible for COVID-19, comes from Yorgo Modis of the University of Cambridge, who works at the Medical Research Council’s Laboratory of Molecular Biology, the UK’s ‘Nobel prize factory’.
His edited responses are in italic to distinguish them from my commentary.
Can we take pictures of viruses?
You can use an electron microscope to image the virus. When we see the virus, we can instantly understand why the coronavirus gets its name – there’s a crown-like haze around the particles, which are spikes that it uses to latch on to human cells.
What is not yet available is a high-resolution 3D reconstruction of an entire, intact SARS-CoV-2 virus.
Viruses are so small that they cannot be seen by a traditional microscope: the wavelength of light is too big for the job.
This new coronavirus measures about 125 nanometers (125 billionths of a metre) in diameter, compared to about 10 micrometers (one millionth of a metre) for an animal cell and 400-600 nanometres for the wavelength of visible light. A human hair is around 80,000- 100,000 nanometers across.
The high voltage electrons in an electron microscope have a wavelength small enough to reveal individual viruses as blurry particles, with their telltale dense interior, lipid (fat) envelope and protein spikes.
Are viruses alive?
Viruses are on the borderline of life, which is a surprisingly slippery concept to define given that we all think we know what life is when we see it.
Each one consists of a package of genetic instructions wrapped in an overcoat of fat and protein, complex molecules made up of strings of amino acids that are the building blocks of living things.
A virus has a single objective: to reproduce.
There is actually a debate about whether viruses are alive (I would say yes).
What is clear is that they are obligate parasites in that they rely on cellular components to replicate – in the case of SARS-CoV-2, the virus relies on our cellular components to multiply.
How do you get a detailed picture of a coronavirus?
You use a computer to combine hundreds of thousands separate electron microscope images of a virus or one of its subcomponents, aligning the images so you get an average structure that is really clear, at a resolution of about one thousandth of a millionth of a metre (one nanometre), which is enough to show the shape of its protein building blocks.
The most modern method, which earned his LMB colleague Richard Henderson a share of a Nobel prize in 2017, is an extension of electron microscopy, called cryogenic electron microscopy, cryo-electron microscopy, or most commonly, cryo-EM.
Older forms of electron microscopy were limited because samples had to be held in a vacuum after coating or staining with metal to make them more visible and withstand the bombardment with electrons.
Using cryo-EM, samples of virus can be maintained in more natural ‘wet’ conditions – a thin layer of vitreous (non-crystalline) ice at liquid nitrogen temperatures – so they do not dry in the vacuum of the microscope and the destructive effect of the electron beam is limited.
Even when aligning the images of thousands of viral particles to produce an average structure, electron microscopes can only resolve features down to around 150,000 Daltons, where a Dalton (named after the pioneering English physical chemist John Dalton) is around the size of a hydrogen atom.
You can just see a viral protein called a polymerase, but not smaller enzymes called proteases, and other components which blend into the background noise.
We have access to one of the highest-resolution cryo-EM instruments in the world (capable of resolving features less than a quarter of a nanometre apart) and aim to exploit that machine to study the virus since this resolution, sufficient to build an atomic model, will be incredibly valuable with the ongoing efforts to find drugs that are more active against SARS-CoV-2.
How do you see the smaller molecular building blocks of viruses?
We also use another technique, called X-ray crystallography, that can reveal even more details of the virus, down to component atoms.
Components of the virus, or even virus fragments attached to a human target protein, are grown into crystals – itself not an easy thing to do, requiring much more virus than electron microscopy – so that millions are arranged in an orderly way for study with intense sources of X-rays.
Because X-rays cannot be focused with lenses, sophisticated software is needed to make a “virtual lens” that can reconstitute images of scattered X rays back into an atomic picture.
There are around 100 different atomic models now (they have been put in the Protein Data Bank) of the building blocks of the virus generated from X-ray crystallography and cryo-electron microscopy.
The images I have seen of the SARS-CoV-2 virus are still fuzzy – why?
The reason is that this coronavirus is not a regular, football-like icosahedral symmetry shape, like so many other viruses, so it is too structurally flexible and heterogeneous to pack into crystals.
This coronavirus has a more fluid envelope of fat and protein with spikes on it, which means that every particle is slightly different and that makes it hard to create an average picture.
The only way to get a high-resolution image is to zoom in on one virus particle, then use a time consuming form of imaging, called cryo-EM tomography.
This method is being used by my colleague John Briggs, working with Andrew Carter, Leo James and Sjors Scheres. They take images of samples of the virus as they are gradually tilted, resulting in a series of 2D images that can be combined to produce a 3D reconstruction – just like tomography in medicine.
Can you follow a virus infecting a human cell?
The first step of infection is for the viral and human cell membranes to fuse, delivering the virus’s genetic instructions inside the cell where they are turned, by human cellular machinery, into new virus particles.
This first step happens fast – on the scale of seconds – once the priming conditions have been met (the right pH and cleavage by one or more proteases) to trigger a change in shape of the Spike protein that starts the fusion process.
Overall, it takes 5-10 minutes for the virus to be internalized from the cell surface. It then takes a couple of hours for the virus’s genetic code to be replicated and for new virus particles to be assembled. New viruses could thus emerge from infected cells within hours (less than a day) of infection.
Can you see what a virus is doing inside human cells?
We have to infer quite a bit about what is going on, for instance from the influence of pH (acidity) in internal structures called endosomes but we can literally drill into cells using a variant of cryo-EM tomography.
You flash freeze an infected cell, then produce cutaways of the cell, shaving down layers of the cell using beams of charged particles called ions until the sample is thin enough to image in an electron microscope.
Ion beam milling is a pretty cool technique in every way and enables you reveal dynamic processes inside a single cell, such as individual coronaviruses in the act of fusing their envelope with the cell’s own envelope.
You won’t ‘see’ atoms but you can see membranes, proteins and make out more detailed structures inside human cells and see how the virus multiplies and self assembles.
Viruses have evolved to hijack cells to make more viruses, no more, no less.
The genetic code of the virus consists of a long, chain-like molecule called RNA, which contains the instructions to make proteins.
Some proteins make up the virus itself, such as the spike, while others help the virus to reproduce and are given arcane names such as NSP7, NSP8 and NSP12, where NSP refers to ‘non-structural protein’.
During an infection, when a human cell is hijacked to make more viruses, the RNA is copied by viral proteins and then proofread by another protein called NSP14. The new copy of RNA is enfolded by viral components made in the cell to create a new virus.
With the help of other coronavirus genes – such as NSP3, NSP4 and NSP6 –newly-minted viruses accumulate in bubbles, called vesicles, that move to the surface of the infected human cell and then spill out, so the new viruses can spread into your lungs, throat, and mouth.
Why is it important to see these details?
By studying each step in the process by which viruses turn our cells into virus factories, scientists get new insights and ammunition for the fight against them.
We now have a really detailed picture of the spike protein, the main feature on the outside of the virus, which consists of two parts, called S1 and S2.
Small proteins, called E and M, are embedded in the viral membrane (they are much smaller than Spike) and another, N, is inside the virus to help package the RNA genome and keep it compact.
With high-resolution structure, such as those from X-ray crystallography and some cryo EM structures, you can design a drug to interfere with a viral protein, just as a lock fits a key.

How complicated is a virus?
While human beings rely on six billion letters of genetic code, three billion from each parent, SARS-CoV-2 relies on just 30,000 ‘letters’ of genetic code.
The order of chemical ‘letters’ in the RNA molecule spells out how to make each of its building blocks, proteins. While humans have at least 20,000 proteins (probably many more), a coronavirus has just 29 proteins.
Turning viral genetic material into a virus is different from the way our cells work. The virus genetic material, an RNA molecule, is first turned into one long ‘polyprotein’ (by contrast, each human gene, roughly speaking makes one protein or protein subunit).
The virus polyprotein is then chopped up into the 29 proteins by enzymes called proteases, some of which are viral – such as one called NSP5 – and some are human proteases already present in infected cells.
My colleagues John Briggs and Sean Munro are investigating the fact that one drug of interest to fight COVID-19 is chloroquine, which affects a small human protein called furin that helps chop up the polyprotein into the proteins that assemble into a new virus.
How is knowledge of the spike protein helping the fight against COVID-19?
My colleague John Briggs is also performing detailed studies of the virus spike protein, which binds to human cells through a docking point, called a receptor, in this case one called ACE2 that responds to a human hormone called angiotensin.
This understanding could help develop drugs, which could work by interfering with the Spike and thus the ability of the virus to infect human cells, but can also help develop tests and vaccines, both of which depend on how immune molecules that are made by an infected person, called antibodies, bind on the spike to neutralise the virus.
Nick Brindle of the University of Leicester is working with my colleague Julian Sale to evolve the region of the ACE2 receptor to which SARS-CoV-2 binds to make a decoy molecule that can bind more effectively to the virus.
With my LMB colleagues – Radu Aricescu, John Briggs, Andrew Carter, Leo James, Jan Löwe (the director of the lab) and Sjors Scheres – we have established ways to produce purified fragments of the spike protein, or whole spike, in the laboratory, and have joined the national COVID-19 Protein Portal to provide these protein reagents free to other labs.
Spike or spike fragments can be injected as a vaccine or used as a diagnostic, to see if antibodies in a pinprick of patient blood react with the virus protein, suggesting that person has encountered the virus before.
How else can this knowledge help drug development?
After the spikes on the surface of the coronavirus have latched on to ACE2 receptors on the surface of the target human cell, the virus enters the cell through endocytosis, a process by which cells normally take in nutrients and other cargo.
My colleagues Leo James, Harvey McMahon and Sean Munro are investigating which of the known endocytosis pathways are exploited by the virus. It is conceivable that a locally applied inhibitor of these pathways could be developed into a drug.
Proteins that carry out chemical reactions – enzymes – are also good targets for drugs as compounds that interfere with the chemical reaction will usually also interfere with viral infection.
Examples of SARS-CoV-2 enzymes are the proteases (NSP3 and NSP5) and the polymerase (NSP12).
How do you figure out how the virus uses human cellular machinery?
We need to understand the metabolic pathways of human cells that the viruses exploit.
One way is to knock out proteins to see what effect that has. Another approach is for us to see which proteins are more or less abundant when virus infects a human cell to map how the virus interacts with cellular pathways.
One key viral protein is called polymerase, which the virus uses to copy its genetic material. My colleagues David Barford, Jan Löwe, Sjors Scheres, and John Sutherland are studying how this important enzyme is disrupted by two potential drugs of interest, Avigan and Remdesivir.
How does SARS-CoV-2 hijack human cells?
This seems to be something to do with a 48 ‘letter’ loop of RNA in the virus genetic code that helps the virus to hijack the cell’s translation machinery (notably huge molecular factories called ribosomes, cellular machines that turn genes into protein and, ultimately, flesh and blood).
Working with Ben Luisi at Cambridge University, Helena Maier at the Pirbright Institute and Bill Scott of the University of California at Santa Cruz, my LMB colleagues Chris Oubridge and Felix Randow are seeking to identify proteins or nucleic acids that bind to this RNA element (this is called RNA interference).
If they are successful, this could in time lead to a drug that is very specific for this virus.
Leo James and Lori Passmore are studying the virus’s N protein, which is thought to protect the virus RNA by linking together to wrap around the RNA, while Venki Ramakrishnan (who won the Nobel prize for his work on the molecular structure of the ribosome) is focusing on another viral protein, NSP1, which helps pirate human cellular machinery to make more virus proteins and prevent it from making antiviral proteins that could stop the virus.
What else is your laboratory doing regarding COVID-19?
We are also looking at a novel way to spot people who have immunity to SARS-CoV-2 by harnessing bacteriophages (or just phages), which are viruses that infect bacteria.
Phil Holliger, working with Willem Ouwehand at the University of Cambridge, is using phages as what they call ‘a self-assembling nanoparticle detection reagent’.
Phages are one of the workhorses of molecular biology and phage technology (for which Greg Winter of the LMB received the Nobel Prize in 2018) allows proteins and peptides (small proteins) to be shown on the surface of the phage particle.
They propose to use phage particles that display COVID-19 viral proteins and peptides as self-assembling nanoparticles that can be simply grown in bacteria and used in a cheap, rapid and easily-scalable phage-based assay for detection and analysis of patient anti-SARS-CoV-2 immune response.
Moreover, Madeline Lancaster and Leo James are also growing nerve cells into ‘mini brains’, called organoids, to see if viruses can invade nervous tissue, following up reports of some patients with COVID-19 experiencing neurological symptoms.
Julian Gough is harnessing his pilot phenotype prediction genome data study to try and identify whether your genotype (genetic make-up) can be used to predict COVID-19 risk factors.
In an earlier blog post, it was noted that there is remarkable diversity in the way people react to the virus.
Gough added: ‘Although the main risk factors in coronavirus infection are age and pre-existing conditions, it is yet to be determined if genetics also has a minor but important role to play … a role that could prove crucial to some people who do not know they are at high risk.
Contributing your genome to medical research is a charitable donation that could have a more profound effect on people’s lives and be even more valuable than a cash donation.’
Do we understand all 29 proteins made by the virus?
There are plenty of things we don’t understand about the COVID-19 virus.
There are some viral proteins that need more study, such as NSP9, which enters the infected cell’s nucleus, and NSP15 that might help clean up viral genetic material so as not to alert the defences of an infected cell, along with a protein called ORF7a which might help viruses escape cells.
We are also still trying to figure out the function of the viral proteins NSP2, NSP11, ORF8 and ORF10.
Many other laboratories around the world are working on the molecular workings of COVID-19 proteins, from the UK’s Diamond Light Source of X-rays to the MRC London Institute of Medical Sciences, University of Texas at Austin,Tsinghua University, China, and the University of Lübeck, Germany.
Stopping the pandemic could rely on this race to visualize SARS-CoV-2 proteins and applying this knowledge in tests, drugs and vaccines.
The latest picture of how far the pandemic has spread can be seen on the Johns Hopkins Coronavirus Resource Center or Robert Koch-Institute. You can check the number of UK COVID-19 lab-confirmed cases and deaths and figures from the Office of National Statistics.
There is more information in my earlier blog posts (including in German by focusTerra, ETH Zürich, with additional information on Switzerland), from the UKRI, the EU, US Centers for Disease Control, WHO, on this COVID-19 portal and Our World in Data.
The Science Museum Group is collecting objects and ephemera to document this public health emergency for future generations.