I talked to John Briggs of the MRC Laboratory of Molecular Biology, Cambridge, one of many labs around the world imaging the SARS-CoV-2 virus at the most intimate, atomic level to devise new ways to combat the pandemic.
His edited responses are in italic to distinguish them from my commentary.
Scientists in the LMB study the virus with X rays, or electron microscopes, notably a method called cryo-electron microscopy, cryo-EM, that, like X rays, is now able to ‘see’ individual atoms due to recent improvements.
The team can also take cryo-EM images of virus samples as they are gradually tilted, resulting in a series of 2D images that can be combined by computer to produce a 3D reconstruction – just like tomography in medicine.

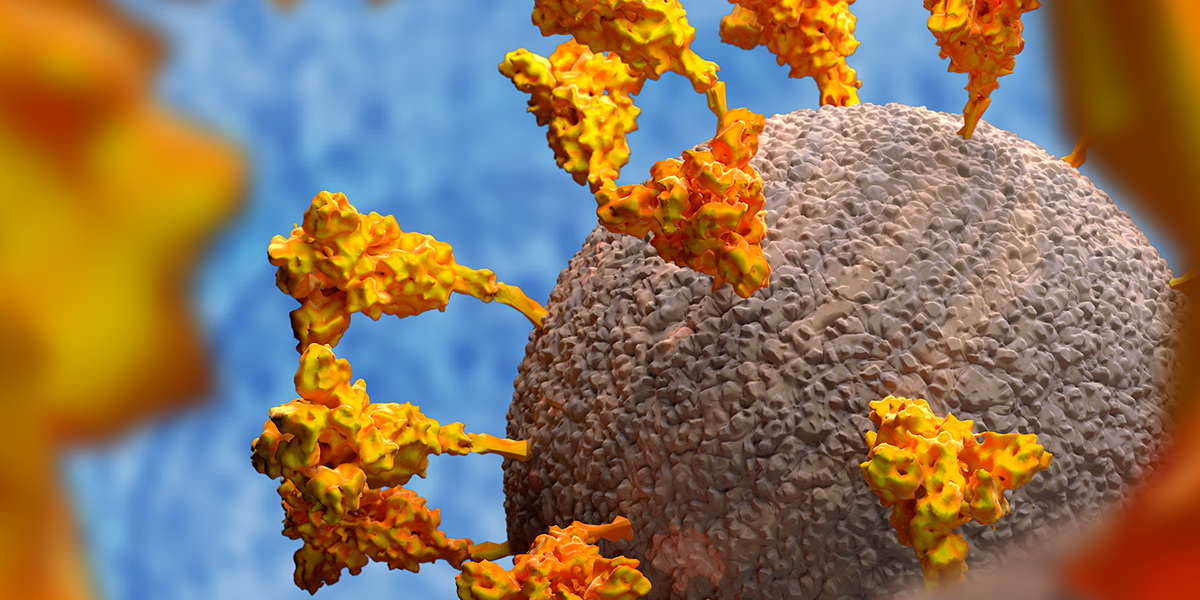
WHAT IS THE SPIKE?

Proteins, complex chemicals, are the building blocks of living things. The spike is one of just 29 proteins described by the virus’s 30,000 letters of genetic code (by comparison, we have six billion).
The spikes are around 25 nanometres long– 25 billionths of a metre – and these are big relative to the virus size compared to some other viruses that we are used to looking at.
When you use an electron microscope to image the virus – which measures 125 billionths of a metre across – you can see why the coronavirus gets its name: there’s a crown-like haze around the particles which consists of the spikes that it uses to latch on to human cells, the first step required for the virus to invade them.
Thanks to research by David Tyrell, head of the Common Cold Unit near Salisbury, and virus-imaging pioneer June Almeida, the first coronavirus was identified and imaged in the 1960s.

WHAT DOES THE SPIKE DO?
The spike – there are between 25 and 40 on the surface of each virus – has evolved to stick to proteins on the surface of many cell types that responds to a human hormone that helps maintain blood pressure called angiotensin – hence their name, angiotensin-converting enzyme 2 (ACE2) receptors. Each spike consists of three copies of the spike protein, S.
HOW DO WE REPRESENT THE SPIKE?
Complicated biological molecules like proteins consist of vast numbers of atoms.
In the case of titin, which makes muscles elastic, its chemical formula is C169,719H270,466N45,688O52,238S911, where C, H, N, O and S refer to atoms of carbon, hydrogen, nitrogen, oxygen and sulphur, respectively.
Here you can see two ways to show the spike protein, either in the form of chains of atoms or by showing the atoms themselves. Vast computer simulations of the protein have been carried out to understand the movement of the spike, such as this one on the Summit supercomputer in Oak Ridge National Laboratory, Tennessee, which included 305 million atoms in the envelope of the virus.


WHAT DOES THE SPIKE LOOK LIKE, UP CLOSE?
The name of this protein is misleading since each spike – which consists of three spike proteins – is not ‘sharp’.
Each spike protein links with two others, forming a tulip-like shape. A stem anchors the proteins to the virus so, when viewed from above, it looks like a flower with three petals.
Nor is the stem rigid, as you would expect of a spike because it is jittering around as a result of thermal energy – the higher the temperature, the more frenetic the molecular movements.
Almost two centuries ago, the Scottish botanist Robert Brown became fascinated by the incessant zigzag motion of fragments in pollen grains, a seemingly obscure phenomenon that would come to be named after him and provided Albert Einstein with proof that atoms exist.
Called ‘Brownian motion’ in his honour, this jiggling caused by heat is central to biology and how complex proteins work. As you read this, the molecular jig goes on within your cells to move your eyes, digest your food and help you to think.
By combining molecular dynamics simulations and cryotomography, Sören von Bülow, Mateusz Sikora and Gerhard Hummer at the Max Planck Institute of Biophysics in Germany identified what they likened to the three joints – ‘hip, knee and ankle’ – that give the stalk its flexibility. Here is a computer simulation of the movement:
HOW DOES THE SPIKE STICK TO HUMAN CELLS?
Each of the three petals of spike protein carries a ‘hook’, called the receptor binding domain (RBD), which sticks to a human cell by binding to the ACE2 receptors on its surface.
There are three copies of the receptor binding domain on the top of this trumpet or tulip shape. They can flip up and down and this movement is stochastic, that is random and caused by heat energy. This is important as the domain must lift up for the binding site to be properly exposed so it can interact with ACE-2. The RBD only sticks out about a billionth of a metre when extended, but the tips of it moves almost four billionths of a metre overall.
WHAT HAPPENS WHEN THE SPIKE LATCHES ON TO A HUMAN CELL?
One, or possibly more, of the receptor binding domains interact with ACE2 molecules on the cell surface. This binding triggers a conformational change – a shape change – within the spike protein.
The inner core of the spike includes fusion peptides – smaller protein components – which can insert into the membrane of the human cell where the ACE-2 was.
When the fusion peptide is inserted, there is a dramatic conformational – shape – change which refolds the entirety of that inner core.
After fusion, the spike becomes longer and thinner (see figure 2b) and then buckles in the middle.

The effect of that refolding is to bring the bit of the spike stuck in the target membrane of a human cell close to the bit which is anchored into the membrane of the virus.
By pulling the two membranes together, it fuses the virus with the cell. When they fuse, the inside of the virus mixes with the inside of the cell.
This enables the viral genetic code to hijack the cell, reprogramming it to become a virus factory with the help of the membranes around the cell’s internal organs, or organelles.
The cell’s own internal membranes are remodelled by the virus and used as assembly sites to bring spike protein together with other viral components to make viral particles.

WHY DOES THE SPIKE HAVE A SUGAR COATING?
Technically, the spike is more than a protein – it’s called a glycoprotein, which means that it has a coat of glycans, sugar-like molecules, on its surface.
Studies, such as this one at the University of California, San Diego, show that the spike protein, including the stalk, is covered with glycan chains to provide a kind of protective coat that hide the spikes from the body’s protective immune system, notably neutralising antibodies.
Rommie Amaro and colleagues in San Diego, Maynooth University, Ireland, and the University of Texas at Austin created computer models, which presented a detailed snapshot of every atom in the spike glycoprotein, revealing regions that weren’t coated by glycans that could be vulnerable to antibodies, especially after the shape change.
You can see the sugar coating of the virus spike protein here.

The glycans are quite flexible, and because of the nature of our methods we only see molecular features that are reasonably stable in position. That means that we see the bits of the glycans that are close to the spike. They help to hide these parts from the immune system. When the RBD at the end of the spike flips up, it seems to lie above the sugar shield

HOW IS THE SPIKE HELPING TO DEVELOP VACCINES?
Vaccines introduce inactivated virus (or parts of the virus) to our immune system to train it to recognise them as an invader so, if ever exposed to the actual virus, the body knows how to fight the infection.
The spike has been central to highly-promising vaccines developed by Pfizer/BioNTech, Moderna and Oxford University.
The first two are RNA vaccines. This kind of vaccine uses genetic information – in the form of RNA – to reprogramme human cells to make the spike protein, which provokes the immune system to respond.
The Oxford vaccine uses viruses themselves as ‘vectors’ to introduce coronavirus proteins to train the body’s immune system. The vaccine consists of a harmless, weakened adenovirus that usually causes the common cold in chimpanzees. The virus has been genetically changed so that it is impossible for it to grow in humans and contains the genetic sequence of the spike protein.
When the vaccine enters cells inside the body, it uses this genetic code to produce the surface spike protein of the coronavirus. This induces an immune response, priming the immune system to attack the coronavirus if it later infects the body.
Most of the vaccine manufacturers have gone for the spike itself, the prefusion form that is on the surface of the virus before it fuses with a human cell.
Because the spike can flip randomly into the long “postfusion” form it adopts after fusing, Moderna and Pfizer use RNAs that change two of the spike protein building blocks, stabilising it into the form it adopts before fusing with human cells. This ‘2P mutation’ was described before this pandemic based on a study on other coronaviruses as a way of stabilising the spike’.
It is a very exciting time now because the data on vaccines suggest they seem to be working. However, there are still open questions about how long-lived the resulting immunity will be, or how robust it will be against strain variations in the longer term.

CAN COVID-19 DRUGS TARGET THE SPIKE?
Understanding the spike can help develop drugs which could work by interfering with the spike and thus hamper the ability of the virus to infect human cells.
Nick Brindle of the University of Leicester is working with Julian Sale at the LMB to evolve the region of the ACE2 receptor to which SARS-CoV-2 binds to make a decoy molecule that can bind more effectively to the virus.
One study used molecular simulations to show that the spike protein adopts an intermediate state before it can dock to the receptor protein on the host cell membrane. This intermediate state can be useful for drug targeting to prevent the spike protein adopting the right shape.
Other groups around the world are probing the binding pocket of the virus, hoping to find a drug that can block the virus from latching onto human cells. One such ‘druggable pocket’ has been discovered by a team headed by Christiane Schaffitzel from Bristol’s School of Biochemistry and Imre Berger from the Max Planck-Bristol Centre for Minimal Biology.
They used electron cryo-microscopy to reveal a small molecule, linoleic acid (a fatty acid we get from our diet), buried in a tailor-made pocket within the spike. Berger said: “Here we have linoleic acid, a molecule which is at the centre of those functions that go haywire in COVID-19 patients, with terrible consequences. And the virus that is causing all this chaos, according to our data, grabs and holds on to exactly this molecule – basically disarming much of the body’s defences.”
CAN WE MAKE SPIKE PROTEIN?
Yes. Many laboratories are making spike protein and fragments including an LMB team – Yorgo Modis, Radu Aricescu, John Briggs, Andrew Carter, Leo James, Jan Löwe (the director of the lab) and Sjors Scheres.
Purified spikes can be used as a diagnostic reagent – for example, ours are being used by collaborators in Addenbrookes in Cambridge to test patients or healthcare workers. They see if antibodies in patient blood react with the virus protein – if they do it suggests that person has encountered the virus before.
We’ve also, like other people, been doing experiments to look at whether different spike proteins stabilised in different ways generate different immune responses. We are also interested in how the spike works with all these confirmational changes that are a little hard to understand.
Different people create very different patterns of antibodies and a lot of those are against the spike of course. Some of those antibody responses are better or worse in terms of patient outcomes. If you start to know whether there are particular patterns which are good, then you could start thinking about whether you could tune the vaccines towards those kinds of immune responses.
Understanding how the spike interacts with an antibody can also allow you to work out the combinations of antibodies that will be able to bind simultaneously for example to different places on the spike because we know from other viruses that combinatorial antibody therapies – a mixture of different types – is more effective.
A lot of researchers are looking for those rare antibodies which are particularly potent and are particularly effective because they act in interesting ways. They could be used as a therapy for people where the immunisation has not worked. If a vaccine is 95% effective that means 5% of people are not protected. It will be a long time before vaccination gives us herd immunity and it will rely on a lot of people taking up the vaccine. For those who are not protected by the vaccine there is still going to be the need for these kinds of therapies.
I think this pandemic has taught us that we should probably have things up our sleeve for future coronaviruses or more general strategies for other potentially pandemic viruses.

CAN YOU USE LAB MADE SPIKE PROTEIN AS A VACCINE?
In principle, yes, though full-length spike protein is hard to produce in large amounts. Novavax has a vaccine in clinical trials using lab made spike protein.
CAN THE SPIKE HELP UNDERSTAND HOW THE VIRUS SPREAD?
The genetic makeup of the virus is being tracked by the COVID-19 Genomics UK Consortium (COG-UK) and, even though the virus does not mutate much, the ancestral form of SARS-CoV-2 that emerged from China has since been largely replaced by strains containing a D614G mutation in the spike.
A study by a Japanese-American team suggests the D614G change in the spike protein, appears to have evolved to enhance transmissibility. Yixuan Hou, Ralph Baric and colleagues did a series of experiments in human cells and animal models to compare the new variant against the ancestral form. The new variant showed an enhanced ability to infect upper airway epithelial cells and to replicate. The good news is that they concluded that the current vaccine approaches should be effective against the D614G strains.
We have put that mutation into the spike, as have many others, to understand what it does to the spike. There is speculation that it might allow the spike to more easily go through the shape changes to expose the receptor binding site and go through fusion. But it may turn out to be more subtle than that, so we are still analysing our data.
I think that any mutation which dramatically changes the spike is likely to render it dysfunctional in some way, so it is likely the mutations which are important are those which either alter the affinity of the spike to receptors or antibodies. These are mutations that are quite subtle in terms of the spike structure and just alter how they bind things – they are what we need to worry about.
D614G is probably one of those mutations, that subtly alter the kind of conformational dynamics of the protein so it still does the same things but some steps in those transitions become a tiny bit faster or a tiny bit slower and that can be enough to have a quite a dramatic effect on the overall efficiency of the life cycle. Anything that completely changes the spike is likely to kill the virus.
Analysing the structure of these spikes could provide clues about how SARS-CoV-2 evolved to infect humans and whether this happened because the virus jumped directly from coronaviruses in bats or via an intermediary species.
Researchers at the Francis Crick Institute in London have compared the structure of the SARS-CoV-2 spike protein and its equivalent in a bat coronavirus, RaTG13s.
While the spikes were over 97% similar, there were significant differences where SARS-CoV-2 binds with ACE2, and at the surfaces that keep the subunits of the spike together. These differences suggest the spike of SARS-CoV-2 is more stable and is able to bind around 1,000 times more tightly to a human cell than this bat virus.

WHAT CAN WE LEARN FROM THE ANTIBODY RESPONSE TO COVID-19?
The authors studied the presence of three types of antibody to COVID-19 in their study: immunoglobulin G (IgG), which is evidence of past infection; IgM, which indicates more recent or current infection, and IgA, which is involved in the mucosal immune response, for example in the nose and respiratory tract.
Of the measured antibody responses, positivity for IgG against the SARS-CoV-2 spike receptor-binding domain was predictive of survival rate.
CAN WE SHOW THE ENTIRE VIRUS ANATOMY?
At the King Abdullah University of Science and Technology, KAUST, in Saudi Arabia, information from electron microscope images and protein databases has been used to created a detailed 3D model of SARS-CoV-2, which can be updated as new data becomes available.
For SARS-CoV-2, they combined information on the shape and size of the virus’s membrane, the spikes, its single RNA strand and the ‘nucleocapsid’ proteins protecting it. “Our visualization presents the latest knowledge about the virus’s mesoscale structure, which is published in Nature, Science and Cell”, said doctoral student Ngan Nguyen.
‘First, our modelling tool gathers important structural aspects from the available imaging evidence, then uses them to form a statistical shape model of the viral particle along with the distribution of proteins on the membrane. For those parts that cannot be seen from images, the rapid modelling system works out structural relationships.
With our tool, we can update the model when the new data about the structure is updated and then regenerate any amount of virus particles as needed.
Our model can be seen used for animations such as this one by Nanographics GmbH.”
WHO WERE THE PIONEERS IN THIS FIELD?
British scientists played a key role in the field of virology and studying the molecular workings of the living world, notably in Cambridge, where the double helix structure of DNA was revealed.
This research continues in many laboratories, notably the Medical Research Council’s Laboratory of Molecular Biology featured here, the UK’s ‘Nobel prize factory’.
The Science Museum Group has many iconic objects from the history of structural biology in its collections and recently hosted an event with the distinguished chemist John Meurig Thomas (who sadly died this month) about the ‘architecture of the invisible’.
HOW CAN I FIND OUT MORE?
The latest picture of how far the pandemic has spread can be seen on the Johns Hopkins Coronavirus Resource Center or Robert Koch-Institute website.
You can check the number of UK COVID-19 lab-confirmed cases and deaths along with figures from the Office of National Statistics.
There is much more information in our Coronavirus blog series (including some in German by focusTerra, ETH Zürich, with additional information on Switzerland), from the UKRI, the EU, US Centers for Disease Control, WHO, on this COVID-19 portal and Our World in Data.
The Science Museum Group is also collecting objects and ephemera to document this health emergency for future generations.